**Notice** As of July 2023, a Convolutional Neural Network that can classify 5 species of Candida with over 90% accuracy was trained. In collaboration with the BioInnovation Laboratory, a white paper is being written for publication. The work below is a demonstration of logistic regression on two species.
What is Candida?
Candida is a genus of over 200 species of yeast. Some of these live naturally in our body, while others are pathogenic and can infect immunocompromised individuals – leading to increased mortality rates. Not only can Candida be dangerous, it commonly takes place in hospitals and is resistance to traditional disinfectant techniques.
Candida identification between species is highly important. This is because, for each species of Candida, a different mixture of medication is required. In addition, species are commonly misidentified and the process requires expensive lab equipment and time.
We seek a new method of diagnosis, that is cost and time efficient. While only require basic lab equipment: A microscope, camera, and computer. With these requirements, countries that do not have access to chemicals, and expensive equipment, will still be able to diagnose the species of Candida and react accordingly.
Data Collection and Analysis
Special thanks to the BioInnovation Laboratory at KSU for providing Candida cultures and an environment for data collection.
Two species of candida were selected: Candida auris and Candida haemulonii.
These two species were selected in particular because they are commonly misidentified between each other.
Cell cultures were grown out for each species, and a collection of 10 pictures were taken for each species. Photographs of each species are shown below. Can you tell the difference by eye? Me either.
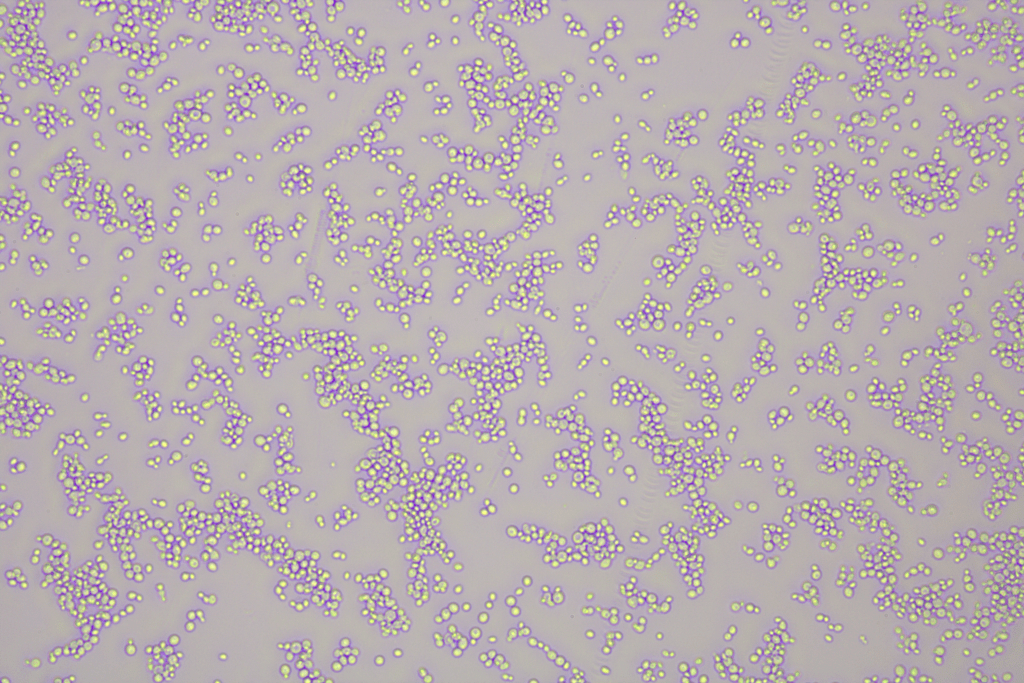
For each image an image analysis software named Fiji/Image J was used to analyze the cell size and output measures into a CSV file
For each image, cells were isolated from one another and the following were outputted for each cell:
- Cell Area
- Circularity (how much like a circle is the cell – from 0 to 1)
- Aspect Ration(how horizontally/vertically stretched is the cell) -defined by [major axis]/[minor axis]
- Solidity (another measure of area) defined by [area]/[convex area]
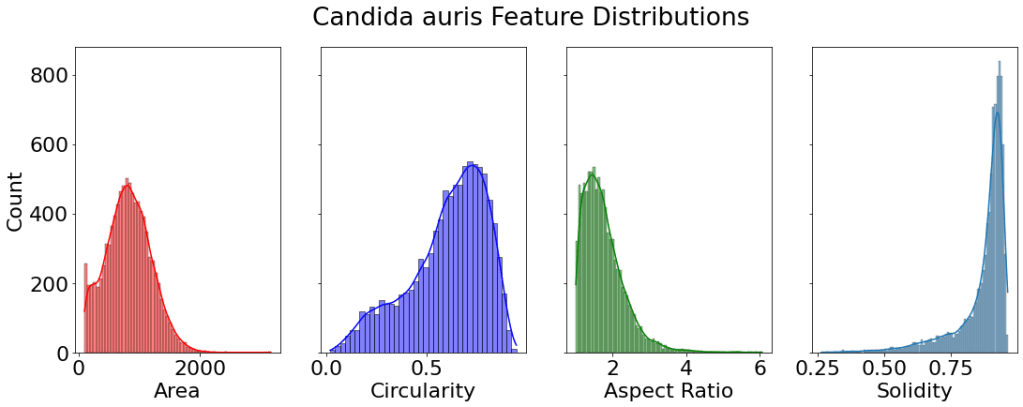
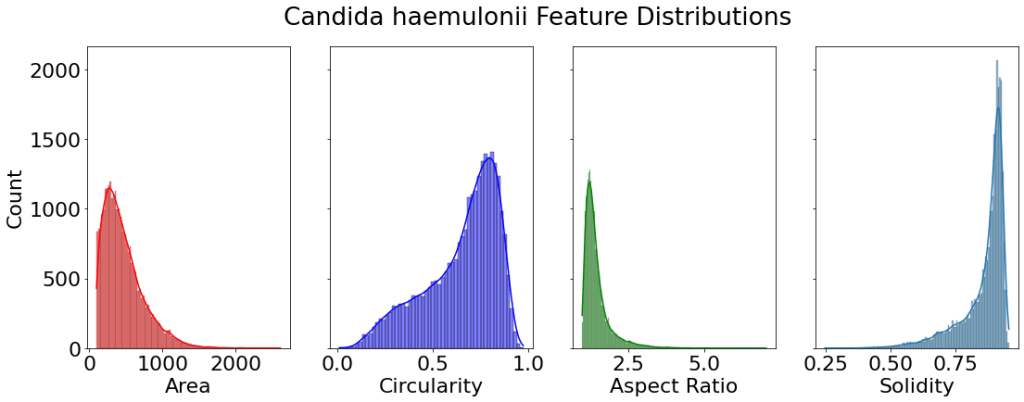
All analysis and figures were made in Python. A two-sample t-test was run on the cell area between species. With a null hypothesis that the two areas are equal. We obtain a p-value of 5.5e-13. Which is much less than our significance level of 0.05. Thus we reject the null hypothesis and conclude there is evidence for the Cell Area of the two species to be different.
Logistic Regression
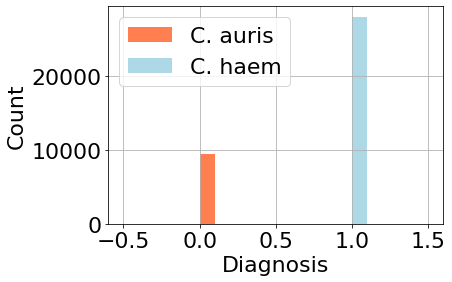
Our data counts were skewed to the Candida haemulonii, due to the fact that this species grows faster than Candida auris. This data imbalance will show in our results.
For training the logistic model: 75% of the data was a training set. While the remaining 25% was used to test.
After the model was trained we obtained the following report:
| Species | Precision | Recall |
| C. auris | 0.73 | 0.44 |
| C. haemulonii | 0.84 | 0.95 |
We see that the model precision is fair. But when classifying Candida auris we have high precision and low recall. This means the model returns relevant results of C. auris but also has a high rate of false negatives when classifying C. auris. Considering our data proportions of the two species, this could be fixed by increasing the amount of data on this species.
Below we plot our Response Operating Characteristic Curve (ROC) and calculate the area under the curve.
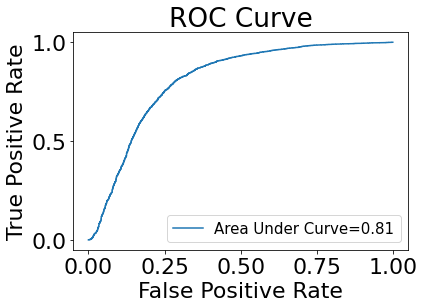
Our Area Under the Curve, which tells us given a random cell, how likely we are to rank a true positive over a false positive. Due to the data-imbalance, our AUC is likely over-estimated.
Improving the model
To get the model in the hands of clinicians we require:
- High Rates of Recall and Precision
- Multispecies Identification
This simple model could increase recall by simply obtaining more pictures. More features could also be generated from the pictures by using the Fiji software, for example Feret’s diameter can be used.
To get to multispecies identification a multivariable logistic regression can be used, but once again requires a larger amount of data.
In the future a Convolution Neural Network could be used, with the input being the images alone. However the advantage to this current model is that it can differentiate given the metrics of a single cell.